Lactate dehydrogenase or LDH (EC 1.1.1.27) is a family of oxidoreductases that catalyze the reversible conversion of pyruvate to lactate, with the concomitant interconversion of NADH and NAD+, which act as cofactors.
They are tetrameric enzymes, where each subunit has catalytic activity. The subunits, encoded by distinct genes, can be assembled in different combination to form isozymes with specific kinetic and regulatory properties.[1]
Lactate dehydrogenase is found in almost all animal tissues, plants, but also in microorganisms. Although it is mostly present in the cytosol, its presence has also been demonstrated in mitochondria, where it catalyzes the oxidation of lactate to pyruvate, and in peroxisomes.[2][3]
In humans, different isoenzymes have preferential tissue localizations, based on the specific metabolic phenotype of the tissue.
Lactate dehydrogenase is a important enzyme in cellular metabolism, as it is involved in energy production from carbohydrates under anaerobic conditions, in the synthesis of glucose from lactate, and utilization of the carbon skeleton of lactate for energy under aerobic conditions.[1][4]
Contents
Genes
In mammals, several genes encode the subunits of lactate dehydrogenase and are designated LDHA, LDHB, LDHC, LDHx, and LDHD.
The first four genes encode enzymes that recognize as substrate the L-isomers of lactic acid, a molecule with a chirality center, the major enantiomeric form of the molecule present in vertebrates, and have NAD as a cofactor, whereas LDHD encodes an enzyme that recognizes as substrate the D-isomer of lactic acid and has FAD as a cofactor.[5][6]
LDHA, located on chromosome 11p15.4, encodes the M subunit, named for its discovery in muscle, whereas LDHB, located on chromosome 12p12.2-p12.1, encodes the H subunit, named for its discovery in heart tissue.[2][7]
LDHC, located on chromosome 11p15.5-p15.3 and probably derived from the duplication of the LDHA, encodes the C subunit.[8]
Finally, LDHx encodes the peroxisomal form of LDH.[2] LDHx is the readthrough form of the LDHB gene; in fact what happens is that when the ribosome reaches the stop codon of the mRNA for the H subunit, reads it as encoding an amino acid. Then the translation proceeds by adding another seven amino acids that constitute the peroxisomal targeting signal by which the proteins are imported into the peroxisome.[3]
Structure
The enzymes belonging to the lactate dehydrogenase family have an oligomeric structure, specifically they are tetramers resulting from the assembly of subunits, of approximately 35 kD, which can be of the same type or two different types. Each subunit has a catalytic site, hence, the tetramer has four active sites. However, the subunits, taken individually, are catalytically inactive.[7][9]
Considering the H and M subunits, their primary structures are approximately 75 percent identical, and consequently, their three-dimensional structure is also very similar, but with small differences at the at the substrate binding site that lead to significant differences in the kinetic properties of the proteins.[1][10] Another consequence of the differences in the primary structure concerns the net surface charge, which is -6 for the M subunit and +1 for the H subunit.[11]
The secondary structure is made up of approximately 40 percent alpha-helices and 23 percent beta sheets, forming beta-alpha-beta structures, with the parallel beta sheets as the main component.[12]
In humans, the prevalent isozymes are those formed by the H and M subunits. The assembly, in different combinations, of the two subunits leads to the formation of five isozymes, namely:
- H4 or LDH-1;
- H3M1 or LDH-2;
- H2M2 or LDH-3;
- H1M3 or LDH-4;
- M4 or LDH-5.[4]
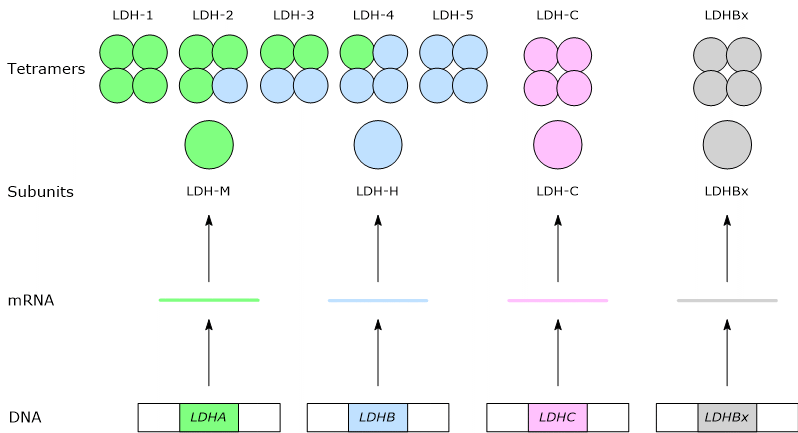 The C subunit forms a sixth isoform, a homotetramer, named as LDH-6 or C4.[8]
The C subunit forms a sixth isoform, a homotetramer, named as LDH-6 or C4.[8]
A seventh isoform derives from the assembly of the LDHx subunit.[3][8]
Reaction
Lactate dehydrogenase catalyzes the reversible conversion of pyruvate to lactate, which are the conjugate base of pyruvic acid and lactic acid, respectively, with the concomitant interconversion of NADH and NAD+. In both cases, the removal of two hydrogen atoms from the reducing agent occurs, followed by the transfer of a hydride ion, namely, a proton and two electrons, to the oxidizing agent, while the remaining proton is released into the aqueous medium as free H+ ion. The enzyme is able to increase the reaction rate by 14 times.[5]
When the reaction proceeds from pyruvate to lactate, the first step is the binding of NADH, the reducing agent, to the enzyme, which is followed by conformational changes that facilitate the formation of hydrogen bonds between specific residues surrounding the active site and the carbonyl carbon of pyruvate, namely C2, and the subsequent interaction between NADH and pyruvate.[13] At this point, a hydride ion is transferred from the nicotinamide ring of NADH to the C2 of pyruvate, which is therefore reduced to lactate. This causes the oxidation of the coenzyme and neutralization of the positive charge carried by the nitrogen of the nicotinamide ring, and reduction of pyruvate to lactate. Therefore, in this case the C2 of pyruvate is reduced from ketone to alcohol.
In the opposite direction, the hydride ion is transferred from the C2 of lactate, which in this case is the reducing agent, to the to the nicotinamide atom C-4. In this case the C2 of lactate is oxidized from alcohol to ketone.[14][15]
Active site
The active site of the H and M subunits has a highly conserved structure in different species, with the same amino acids participating in the reaction.[16] His-193, which is found near the binding site for the coenzyme, is among these, not only for human lactate dehydrogenase, but also for that of many other species.
However, the small differences in their primary structure lead to different biochemical properties. Among the differences, one is crucial for the catalytic properties: an alanine of the M subunit is replaced by a glutamine in the H subunit, namely, a non-polar amino acid is replaced by a polar one.[11] Below is a brief overview of the catalytic differences between the two subunits.
The H subunit binds substrate faster than the M subunit, but has about five times less catalytic activity than the M subunit.
The M subunit has a higher affinity for pyruvate, thus favoring the formation of lactate and NAD+, and is not inhibited by high concentrations of pyruvate. Conversely, the H subunit has a higher affinity for lactate, which favors the formation of pyruvate and NADH, and is inhibited by high concentrations of pyruvate.[17][18]
From all this it follows that the kinetic/catalytic properties of the different isozymes depend on the prevalence of one of the two subunits.[19]
Regulation by pyruvate and lactate
Lactate dehydrogenase isozymes are subject to inhibition by pyruvate and lactate.
The isozymes where the H subunit predominates are inhibited by lower pyruvate concentrations than those where the M subunit predominates. For example, H4 is inhibited by pyruvate concentrations of about 0.2 mM, while M4 is weakly inhibited by pyruvate concentrations up to 5 mM.
Conversely, H4 is inhibited by lactate concentrations greater than 20-40 mM, whereas M4 is inhibited to a lesser extent by high lactate concentrations.[20]
Tissue distribution
In humans, the different lactate dehydrogenase isozymes have preferential tissue localizations, which generally reflect the metabolic phenotype of the tissue.
In fact, tissues with a predominantly or exclusively aerobic metabolic phenotype, such as the heart, produce mainly isozymes in which the H subunit predominate.[21]
By contrast, tissues where anaerobic metabolism is important, such as muscle fibers during vigorous exercise, or hypoxic regions in solid tumors, produce mainly isozymes in which the M subunit predominate.[5]
However, there are exceptions, such as the liver, an organ with an aerobic metabolic phenotype where LDH-5 predominates, but where the oxidation of lactate to pyruvate, which can also be considered as a part of the hepatic branch of the Cori cycle, is favored by the low NADH/NAD+ ratio in the hepatocyte cytosol.[13]
Below is a brief overview.
- LDH-1 or H4: cardiac muscle, kidney and red blood cells.
- LDH-2 or M1H3 has a distribution similar to LDH-1.
- LDH-3 or M2H2: spleen, brain, white blood cells, kidney and lung.
- LDH-4 or M3H1: spleen, lung, skeletal muscle, red blood cells and kidney.
- LDH-5 or M4: liver, skeletal muscle and lung.
- LDH-6: sperm and testis.
In individual organs, LDH-1 and LDH-2 predominates in the heart, kidney and red blood cells, LDH-3 in the lung, LDH-4 and LDH-5 in skeletal muscle and LDH-5 in the liver.
The isozymes present in serum, whose dosage is used for diagnostic purposes, are always of tissue origin.[5]
Finally, in the same tissue/organ, different isozymes can be present in significant quantities in different cell types, as for example occurs in skeletal muscle, in the brain, but also in solid tumors.
Role
Lactate dehydrogenase is involved both in the synthesis and utilization/removal of lactate.[4]
Under hypoxic conditions, the cell obtains ATP from the anaerobic oxidation of glucose. Through the glycolytic pathway, the monosaccharide is oxidized to two molecules of pyruvate, yielding 2 ATP and two NADH. However, for glycolysis to proceed, the NADH produced must be reoxidized to NAD+, as the oxidized coenzyme is involved in the reaction catalyzed by glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12). As the pyruvate dehydrogenase complex is inhibited and oxidative phosphorylation is blocked, the oxidation of NADH occurs in the reaction catalyzed by lactate dehydrogenase, which then allows glycolysis to proceed even in hypoxic conditions. This metabolic pathway produces lactate from glucose, and is known as lactic acid fermentation.[13][22]
Lactic acid is often considered an end product of glucose metabolism, and its accumulation in the body is harmful as it may potentially causelactic acidosis.[22] Therefore, it must be rapidly removed from tissues and circulation. Considering for example the lactose produced by red blood cells or muscle fibers working in under hypoxic conditions, such as during vigorous exercise, or continuously by the red blood cell, through bloodstream, it can reach, among others, the liver and the heart. In the liver, although LDH-5 is the major isozyme, the low cytosolic NADH/NAD+ ratio shifts the equilibrium of the reaction toward pyruvate synthesis, which can be used for energy or enter into gluconeogenesis.[23] In the cardiomyocyte LDH-1 oxidizes lactate to pyruvate, that will be used for energy.
References
- ^ a b c Berg J.M., Tymoczko J.L., and Stryer L. Biochemistry. 5th Edition. W. H. Freeman and Company, 2002
- ^ a b c Markert C.L., Shaklee J.B., Whitt G.S. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science 1975;189(4197):102-14. doi:10.1126/science.1138367
- ^ a b c Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gärtner J., Thoms S. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife 2014;3:e03640. doi:10.7554/eLife.03640
- ^ a b c Laughton J.D., Charnay Y., Belloir B., Pellerin L., Magistretti P.J., Bouras C. Differential messenger RNA distribution of lactate dehydrogenase LDH-1 and LDH-5 isoforms in the rat brain. Neuroscience 2000;96(3):619-25. doi:10.1016/s0306-4522(99)00580-1
- ^ a b c d Farhana A., Lappin S.L. Biochemistry, lactate dehydrogenase. [Updated 2023 May 1]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK557536/
- ^ Jin S., Chen X., Yang J., Ding J. Lactate dehydrogenase D is a general dehydrogenase for D-2-hydroxyacids and is associated with D-lactic acidosis. Nat Commun 2023;14(1):6638. doi:10.1038/s41467-023-42456-3
- ^ a b Krieg A.F., Rosenblum L.J., Henry J.B. Lactate dehydrogenase isozymes a comparison of pyruvate-to-lactate and lactate-to-pyruvate assays. Clin Chem 1967;13(3):196-203. doi:10.1093/clinchem/13.3.196
- ^ a b c Zinkham W.H., Blanco A., Clowry L.J. Jr. An unusual isozyme of lactic dehydrogenase in mature testes: localization, ontogeny, and kinetic properties. Ann N Y Acad Sci 1964;121:571-88. doi:10.1111/j.1749-6632.1964.tb14227.x
- ^ Palmer T., Bonner P.L.. Enzymes – Monomeric and oligomeric enzymes. 2nd Edition, Woodhead Publishing, 2011. doi:https://doi.org/10.1533/9780857099921.1.76
- ^ Bellamacina C.R. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J 1996;10(11):1257-69. doi:10.1096/fasebj.10.11.8836039
- ^ a b Read J.A., Winter V.J., Eszes C.M., Sessions R.B., Brady R.L. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins 2001;43(2):175-85.
- ^ Auerbach G., Ostendorp R., Prade L., Korndörfer I., Dams T., Huber R., Jaenicke R. Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization. Structure 1998;6(6):769-81. doi:10.1016/s0969-2126(98)00078-1
- ^ a b c Forkasiewicz A., Dorociak M., Stach K., Szelachowski P., Tabola R., Augoff K. The usefulness of lactate dehydrogenase measurements in current oncological practice. Cell Mol Biol Lett 2020;25:35. doi:10.1186/s11658-020-00228-7
- ^ Moran L.A., Horton H.R., Scrimgeour K.G., Perry M.D. Principles of Biochemistry. 5th Edition. Pearson, 2012
- ^ Nelson D.L., M. M. Cox M.M. Lehninger. Principles of biochemistry. 6th Edition. W.H. Freeman and Company, 2012
- ^ Holmes R.S., Goldberg E. Computational analyses of mammalian lactate dehydrogenases: human, mouse, opossum and platypus LDHs. Comput Biol Chem 2009;33(5):379-85. doi:10.1016/j.compbiolchem.2009.07.006
- ^ Feng Y., Xiong Y., Qiao T., Li X., Jia L., Han Y. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med 2018;7(12):6124-6136. doi:10.1002/cam4.1820
- ^ Gray L.R., Tompkins S.C., Taylor E.B. Regulation of pyruvate metabolism and human disease. Cell Mol Life Sci 2014;71(14):2577-604. doi:10.1007/s00018-013-1539-2
- ^ Rogatzki M.J., Ferguson B.S., Goodwin M.L., Gladden L.B. Lactate is always the end product of glycolysis. Front Neurosci 2015;9:22. doi:10.3389/fnins.2015.00022
- ^ Stambaugh R., Post D. Substrate and product inhibition of rabbit muscle lactic dehydrogenase heart (H4) and muscle (M4) isozymes. J Biol Chem 1966;241(7):1462-7. doi:10.1016/S0021-9258(18)96733-5
- ^ Wroblewski F, Gregory KF. Lactic dehydrogenase isozymes and their distribution in normal tissues and plasma and in disease states. Ann N Y Acad Sci 1961;94:912-32. doi:10.1111/j.1749-6632.1961.tb35584.x
- ^ a b Li X., Yang Y., Zhang B., Lin X., Fu X., An Y., Zou Y., Wang J.X., Wang Z., Yu T. Lactate metabolism in human health and disease. Signal Transduct Target Ther 2022;7(1):305. doi:10.1038/s41392-022-01151-3
- ^ Gleeson T.T. Post-exercise lactate metabolism: a comparative review of sites, pathways, and regulation. Annu Rev Physiol 1996;58:565-81. doi:10.1146/annurev.ph.58.030196.003025