Animal, vegetable, bacterial, and viral proteins are large linear polymers made up of hundreds and even thousands of subunits called amino acids.
The amino acid sequence, which is unique and genetically encoded, is called the polypeptide chain or primary structure. In the polypeptide chain, the amino acids are linked in series by covalent bonds called peptide bonds.
The primary structure largely determines what will be the three-dimensional structure, or conformation, of the protein.
Proteins are a large and diverse class of molecules, they are present in all living organisms, in all compartments of the cell, and have very different structures, even within the same cell type, where we can find hundreds or even thousands of different types, each of which perform a different function.
The great variety of functions that are able to perform derives from the possibility of the polypeptide chain to fold into specific three-dimensional structures, which also provide the ability to bind different molecules. It can therefore be said that proteins are the tool through which genetic information is expressed.
Contents
The discovery
As proteins are generally easier to isolate than lipids, nucleic acids, and polysaccharides, their study preceded that of other biomolecules, and can be traced back to the works on the chemical composition of albumins conducted by Jöns Jacob Berzelius, considered one of the fathers of modern chemistry, and Gerardus Johannes Mulder, in 1839. By comparison, the role of nucleic acids in the transmission and expression of genetic information came to light in the 1940s, and their catalytic role only in the 1980s, whereas the role of lipids in biological membranes in the 1960s.
The term protein derives from the Greek word proteios, that means primary or preeminent, and was first suggested by Berzelius to Mulder. In fact, Berzelius believed that proteins could be the most important biological substances.
Structure
Like other biological macromolecules, proteins are made up of many small organic molecules, namely, amino acids.
About 20 different amino acids have been identified which, according to Fischer-Rosanoff convention, are present almost exclusively in the L form. Occasionally, D-amino acids have been found in certain bacterial proteins.
L-amino acids are bifunctional organic compounds as they contain both a carboxyl group and an amino group attached to a central carbon atom, known as the alpha-carbon, and for this reason they are also called L-alpha-amino acids. Each amino acid is characterized by a side chain, known as an R group, that, like the carboxyl group and the amino group, is attached to the alpha carbon. R group is responsible for the chemical properties of the amino acid, as it has a variety of sizes, charge, shapes, and reactivities.
During protein synthesis, the carboxyl group of one amino acid is covalently linked to the amino group of the incoming amino acid via a condensation reaction, which is catalyzed by specific enzymes, namely, proteins with catalytic activity. During the reaction, a water molecule is released and a peptide bond is formed. Peptide bond is rigid, planar and very stable bond; indeed, at physiological pH, in the absence of external interventions, its life is about 1,100 years. A linear polypeptide chain is formed by end-to-end bonds between adjacent amino acids.
In the description of how polypeptide chains fold into their three-dimensional structures, it is helpful to distinguish different levels of organization, namely, primary, secondary, and supersecondary structures, domains, and, tertiary and quaternary structures.
Note: Fischer-Rosanoff convention is no longer used, except for carbohydrates and amino acids, replaced by the RS system, which allows to name unambiguously molecules with chirality centers.
Protein primary structure
Bovine insulin was the first protein whose primary structure was determined, thanks to the work of Frederick Sanger.
The primary structure is the amino acid sequence of the protein, its lowest level of organization, and, as mentioned, it is unique, genetically determined, and responsible for the three-dimensional structure and function of the protein.
It can consist of 40 to over 4,000 amino acid residues.
The polypeptide chain has polarity as its two ends are different: one has a free amino group and is called NH2-terminus or amino-terminus, the other a free carboxyl group, and is called COOH-terminus or carboxyl-terminus. The two ends of the polypeptide chain are also known as N-terminal end and C-terminal end, to distinguish them from the carboxyl and amino groups present in the R-groups. By convention, the N-terminal end is taken as the beginning of the amino acid chain, and is written on the left.
The primary structure is also interesting because, by comparing that of a same protein present in different species, it is possible to identify the variations that the corresponding gene has undergone, which are an indicator of the divergence of the species in the course of evolution.
The terms dipeptide, tripeptide, oligopeptide and polypeptide are used to indicate chains of different lengths, respectively consist of 2, 3, less than 50, and more than 50 amino acids.
Protein secondary structure
The discovery of the secondary structure of proteins is due to the work of Linus Pauling and Robert Corey, which proposed two structures called alpha-helix and beta-sheet structure or beta-pleated sheet.
Protein secondary structure results from the formation of hydrogen bonds between contiguous parts of the polypeptide chain having particular amino acid sequences. Therefore, it describes the arrangement in space of amino acids not far apart along the primary structure.
In addition to alpha-helix and beta-sheet structure, others have been identified such as beta-turns, gamma turns, and omega loops, all belonging to the group called reverse turns. These structures are often found where the polypeptide chain reverses direction, and are generally located on the surface of the molecule.
About 32-38 percent of the amino acids in globular proteins are found within alpha-helix structures.
The structures following the secondary one are present only in globular proteins.
Supersecondary structures or motifs
They are combination of secondary structures which form a region of the molecule with a characteristic three-dimensional structure and topology. The supersecondary structures are connected to each other by loop regions with undefined structure.
Among the most common motifs are the alpha-alpha-corner, beta-beta-corner, beta-beta-hairpin, beta-alpha-beta-motif, the latter often present in proteins that bind RNA or DNA, and the 3beta-corner.
Domains
They are globular regions that result from the combination of motifs that fold independently from the rest of the polypeptide chain to give a stable structure.
They consist of 40-400 amino acids, except the motor and kinase domains which are made up of a much larger number of amino acids.
The domains have been classified into three main groups, on the basis of secondary structures and motifs present:
- alpha-domains;
- beta-domains;
- alpha/beta-domains.
More than 1,000 domain families have been identified, and the members of each family are called homologues.
Very often, each domain has a specific function, that is, it is a functional unit of the protein in which it is contained.
Proteins can consist of a single domain, the smaller ones, or of multiple domains. For example, chymotrypsin (EC 3.4. 21.1), an enzyme involved in protein digestion, consists of a single domain, while papain (EC 3.4.22.2) is composed of two domains.
Protein tertiary structure
The tertiary structure, also called the native structure, is the three-dimensional structure of the proteins, and is their biologically active form.
The first protein whose tertiary structure was determined was myoglobin, thanks to the work of John Kendrew.
Polypeptide chains are not rigid structures and spontaneously fold up into distinct three-dimensional structures that are largely determined by their amino acid sequence. The folding of the primary structure allows the transition from the one-dimensional world of the primary structure to the three-dimensional one of the protein.
In this type of structure, the folding of the polypeptide chain causes distant amino acids to find themselves close together, therefore it concerns the three-dimensional arrangement of amino acids far from each other in the primary structure.
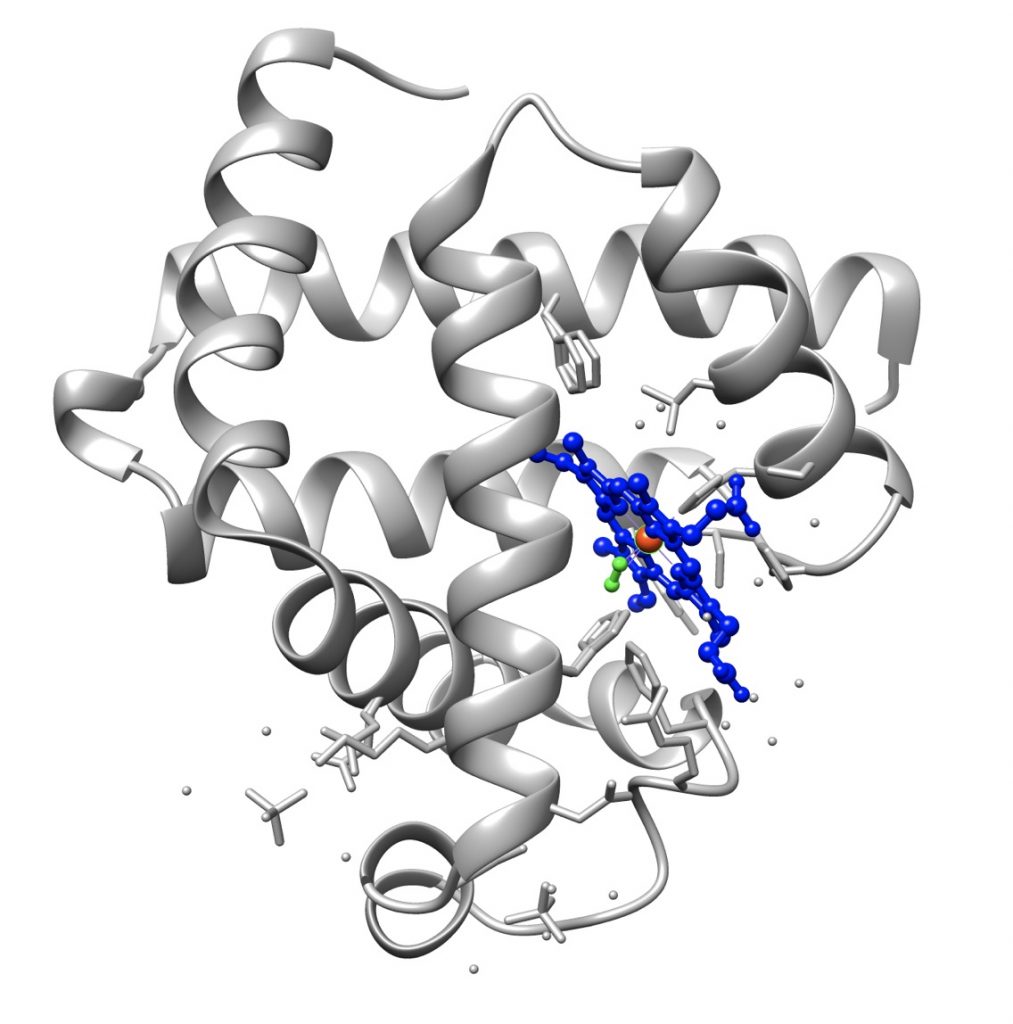
The tertiary structure of the proteins, especially of proteins consisting of more than 200 amino acids, is formed by several domains joined by short polypeptide segments. It is often stabilized by disulfide bridges between cysteine residues, bridges which are formed after the molecule has attained the native conformation.
Not all globular proteins have a tertiary structure. An example are milk caseins, whose polypeptide chain assumes a disordered three-dimensional conformation, also known as random coiled structure. The disordered structure makes them highly susceptible to the action of intestinal proteases, and therefore to the release of the constituent amino acids. Another example of random coiled protein is elastin, one of the most abundant proteins in the body.
Protein quaternary structure
This level of structural organization describes how two or more polypeptide chains associate to form a single protein structure. Therefore, it refers to the spatial arrangement of the individual chains and the nature of the forces that bind them, such as:
- the hydrophobic effect, which is also the main driving force for protein folding;
- hydrogen bonds;
- van deer Waals interactions;
- ionic interactions;
- covalent cross-links.
The resulting structure is called oligomer or oligomeric protein, and the constituent polypeptides, which may be identical or different, monomers or simply subunits.
In general, most intracellular proteins are oligomers, while most extracellular proteins are not. An example of a protein with a quaternary structure is hemoglobin.
This level of structure is obviously absent in globular proteins consisting of a single polypeptide chain, that is, in monomeric proteins.
Proteins can also interact each other to form macromolecular machines in which, acting in a synergistic way, they perform functions that they would not be able to accomplish alone. An example are the multienzyme complexes, such as the pyruvate dehydrogenase complex.
Functions
Proteins are the most versatile macromolecules present in living organisms, and play a central role in virtually all cell structures and functions, such as:
- chemical reactions;
- oxygen transport;
- immune response;
- control of growth and differentiation;
- nerve transmission;
- storage;
- mechanical support;
- movement.
Furthermore, proteins are involved in the digestive processes that take place in the gastrointestinal tract. In fact, the macronutrients, lipids, carbohydrates and proteins, in order to be absorbed must be hydrolyzed to fatty acids, cholesterol, and glycerol, monosaccharides, and amino acids, respectively. These reactions, in the course of protein and carbohydrate digestion, as well as of lipid digestion, are catalyzed by specific enzymes, such as alpha-amylase (EC 3.2.1.1) for hydrolysis of the α-(1→4) glycosidic bonds of amylose and amylopectin, the two polysaccharides that form starch granules.
Note that a method for protein classification bases on their biological functions.
References
- Berg J.M., Tymoczko J.L., and Stryer L. Biochemistry. 5th Edition. W. H. Freeman and Company, 2002
- Fang C., Shang Y., Xu D. Improving protein gamma-turn prediction using inception capsule networks. Sci Rep 2018;8(1):15741. doi:10.1038/s41598-018-34114-2
- Garrett R.H., Grisham C.M. Biochemistry. 4th Edition. Brooks/Cole, Cengage Learning, 2010
- Lodish H., Berk A., Zipursky S.L., et al. Molecular cell biology. 4th edition. New York: W. H. Freeman; 2000. Section 3.1, Hierarchical Structure of Proteins.
- Kessel A., Ben-Tal N. Introduction to proteins: structure, function, and motion. CRC Press, 2011. doi:10.1002/cbic.201100254
- Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 2013;35(12):1050-5. doi:10.1002/bies.201300066
- Moran L.A., Horton H.R., Scrimgeour K.G., Perry M.D. Principles of Biochemistry. 5th Edition. Pearson, 2012
- Nelson D.L., Cox M.M. Lehninger. Principles of biochemistry. 6th Edition. W.H. Freeman and Company, 2012
- Rudnev V.R., Kulikova L.I., Nikolsky K.S., Malsagova K.A., Kopylov A.T., Kaysheva A.L. Current approaches in supersecondary structures investigation. Int J Mol Sci 2021;22(21):11879. doi:10.3390/ijms222111879
- Stipanuk M.H., Caudill M.A. Biochemical, physiological, and molecular aspects of human nutrition. 3rd Edition. Elsevier health sciences, 2012
- Voet D. and Voet J.D. Biochemistry. 4th Edition. John Wiley J. & Sons, Inc. 2011